Biography
I am a computational biologist at the University of Oxford.
My research interests in computational biology include software engineering best practices, DevOps, single-cell genomics, and interactive data visualization. I particularly enjoy using and contributing R packages part of the Bioconductor project. A list of software packages that I maintain or contributed to is available on the Software page.
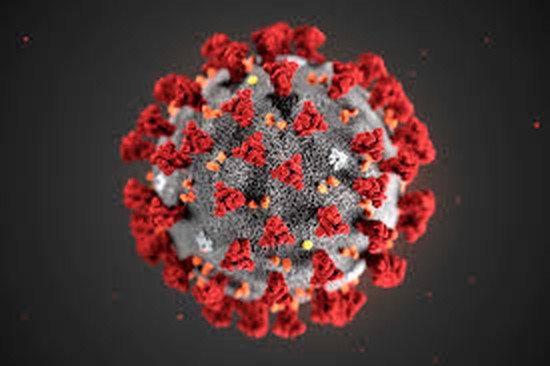
My academic research primarily explores the host immune response to infectious diseases, inflammation, and self-antigens.
Interests
- Computational Biology
- Software Development
- Bioconductor packages
- Single-cell genomics
Education
-
PhD in sa Infection Biology, 2015
United International University
-
MSc in Biological Engineering, 2011
Polytech Nice - Université Nice Sophia Antipolis
-
BSc in Biology, Chemistry, Physics and Earth Sciences, 2008
CPGE BCPST Véto - Lycée Jean Rostand